

- Home
- Companies
- Microbial Insights, Inc.
- Products
- Census - Model (qPCR) ER - Chlorinated ...
Census - Model (qPCR) ER -Chlorinated Ethenes Remediation
CENSUS qPCR (quantitative polymerase chain reaction) is a DNA based method to accurately quantify specific microorganisms (e.g. Dehalococcoides, Dehalobacter) and functional genes (e.g., vinyl chloride reductase, anaerobic benzene carboxylase) responsible for biodegradation of contaminants.
ACCURATE
Direct analysis of sample DNA removes the need to grow the bacteria, thus eliminating biases associated with traditional approaches (e.g., plate counts and MPNs).
QUANTITATIVE
Absolute quantification of the concentrations of specific microorganisms and functional genes encoding enzymes responsible for contaminant biodegradation gives site managers a direct line of evidence to evaluate remediation options and monitor remedy performance. Results reported as cells/mL, cells/g, etc.
INFORMATIVE
Is that a low, medium, or high concentration of contaminant degraders? With the MI Database, clients can retrieve percentile rankings of their CENSUS ® qPCR results to answer that question based on the tens of thousands of samples MI has received from sites around the world.
SENSITIVE
Practical Detection Limits (PDL) are as low as 100 cells per sample with a dynamic range over seven orders of magnitude. Low detection limits are particularly important when evaluating whether bioaugmentation is needed or an unnecessary expense.
SPECIFIC
Target specific bacterial groups (e.g., Dehalococcoides, Dehalobacter, Dehalogenimonas) and functional genes (e.g. vinyl chloride reductases, anaerobic benzene carboxylase) responsible for contaminant biodegradation.
COST EFFECTIVE
Fast turnaround time (7-10 days), with rush service available, so you can make decisions and take action quickly. CENSUS® qPCR is inexpensive and ultimately saves money by allowing site managers to make more informed decisions.
FLEXIBLE
Analysis can be performed on almost any type of sample (water, soil, sediments, Bio-Traps®, and others).
CUSTOMIZABLE
Assays are available for quantification of many microorganisms and functional genes responsible for biodegradation of a broad spectrum of contaminants. Custom assays can also be developed to fit your needs.
The most direct avenue to evaluate biodegradation as a treatment mechanism is to directly quantify the microorganisms or biological processes responsible for biodegradation of the contaminants of concern. Following is a breakdown of the CENSUS targets available for evaluating biodegradation of chlorinated ethenes.
CENSUS Targets for Reductive Dechlorination (Analysis of Dehalococcoides, Dehalobacter, Dehalogenimonas and other bacteria capable of reductive dechlorination)
Under anaerobic conditions, PCE can be sequentially dehalogenated through TCE, cis-dichloroethene (cis-DCE), and vinyl chloride (VC) to ethene via microbially mediated reductive dechlorination. Because ethene is an innocuous end product, reductive dechlorination is an attractive treatment mechanism for PCE/TCE-impacted sites.
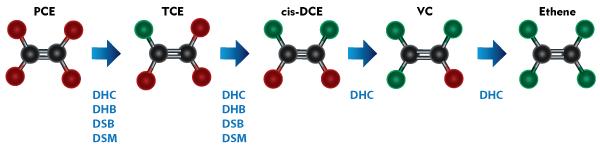
The following table describes the individual CENSUS targets, their importance in evaluating reductive dechlorination as a treatment mechanism, and provides guidelines for integrating CENSUS results into routine groundwater monitoring for common corrective actions.
| Target | MI Code | Relevance / Data Interpretation |
| Dehalococcoides | DHC | Only known group of bacteria capable of complete dechlorination of PCE and/or TCE to ethene. Absence of Dehalococcoides suggests dechlorination of DCE and VC is improbable and accumulation of daughter products is likely. The presence of Dehalococcoides even in low copy numbers indicates the potential for complete reductive dechlorination. Higher copy numbers and the presence of daughter products suggest that dechlorination may be occurring. |
| Dehalococcoides Functional Genes | TCE
VCR BVC |
Functional genes encoding reductive dehalogenases for TCE and VC. Presence of TCE reductase indicates the ability to reduce TCE to DCE and VC. Presence of VC reductases (VCR and BVC) indicates the potential for reductive dechlorination of VC to ethene. Absence of VC reductases suggests that VC may accumulate. |
| Dehalobacter | DHB | Capable of dechlorination of PCE and TCE to cis-DCE but best known for utilizing chlorinated ethanes including TCA and DCA isomers, which are common co-contaminants at PCE/TCE-impacted sites. |
| Desulfuromonas | DSM | Capable of dechlorination of PCE and TCE to cis-DCE using acetate as an electron donor. |
| Desulfitobacterium | DSB | Capable of dechlorination of PCE and TCE to cis-DCE. |
| Total bacteria | EBAC | Index of total bacterial biomass. Domain level. |
| Methanogens | MGN | Methanogens utilize hydrogen and carbon dioxide to produce methane. Compete with dechlorinating bacteria for available hydrogen. |
| Sulfate Reducing Bacteria | APS |
The assay targets a gene involved in sulfate reduction. As with methanogens, SRBs can compete with dechlorinating bacteria for available hydrogen. |
CENSUS Targets for Cometabolism of Chlorinated Ethenes (Methanotrophs and other bacteria capable of co-metabolism)
Under aerobic conditions, several different types of bacteria including methane-oxidizing bacteria (methanotrophs), ammonia-oxidizing bacteria, and some toluene/phenol-utilizing bacteria can cometabolize or co-oxidize trichloroethene (TCE), dichloroethene, and vinyl chloride (VC). In general, cometabolism of chlorinated ethenes is mediated by monooxygenase enzymes with “relaxed” specificity that oxidize a primary (growth supporting) substrate and co-oxidize the chlorinated compound. In the presence of methane, for example, methanotrophs produce methane monooxygenases which oxidize methane to methanol and can also co-oxidize TCE.
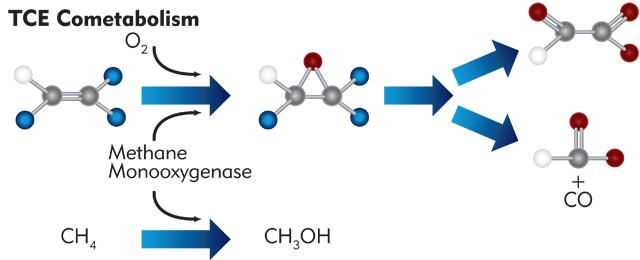
The following table describes individual CENSUS targets and their importance in evaluating aerobic cometabolism as a treatment mechanism, and provides guidelines for integrating CENSUS results into routine groundwater monitoring for common corrective actions.
| Target | MI Code | Relevance / Data Interpretation |
| Methanotrophs | MOB | Targets two types of methane oxidizing bacteria (methanotrophs). Indicates the potential for cometabolic oxidation of TCE. |
| Soluble Methane Monooxygenase | sMMO | Targets the soluble methane monooxygenase gene. Soluble methane monooxygenases are generally believed to support faster cometabolism of TCE. |
| Propane Monooxygenase | PPO | Propane can be added as a primary substrate to promote cometabolic oxidation of TCE. |
| Ring Hydroxylating Toluene Monooxygenase | RDEG | Applicable at mixed waste sites where BTEX and TCE are co-contaminants. When expressed, toluene monooxygenases are capable of cometabolism of TCE. |
| Ring Hydroxylating Toluene Monooxygenase | RMO | Applicable at mixed waste sites where BTEX and TCE are co-contaminants. When expressed, toluene monooxygenases are capable of cometabolism of TCE. |
| Butane Monooxygenase | BMO | Targets the butane monooxygenase gene. |
| Ethene Monooxygenase | ETN | Enumerates key functional genes (etnC and etnE) involved in ethene utilization and vinyl chloride (co)metabolism. |
