

EpiCypher

SNAP-CUTANA - K-MetStat Panel
FromEpiCypher
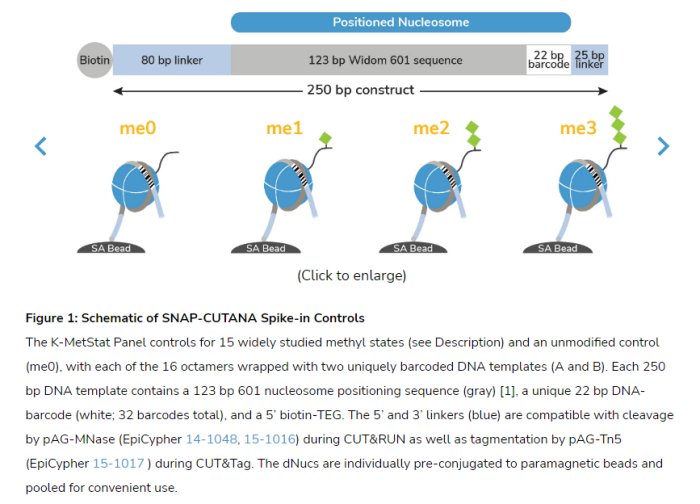
The SNAP-CUTANA K-MetStat Panel of spike-in controls for CUT&RUN and CUT&Tag offers an all-in-one solution to determine antibody specificity for histone posttranslational modifications (PTMs), monitor assay success, and normalize data for quantitative chromatin mapping. The panel contains designer nucleosomes (dNucs) representing 16 different K-methyl PTM states: mono-, di-, and trimethylation at H3K4, H3K9, H3K27, H3K36, & H4K20, as well as unmodified control (Figure 1). Each PTM is represented by two unique DNA-barcoded templates (A and B, for an internal technical replicate). Each dNuc is individually conjugated to paramagnetic beads and pooled into a single panel for convenient one-step spike-in to CUT&RUN and CUT&Tag experiments. The panel is added to samples alongside ConA-immobilized cells prior to the addition of an antibody targeting a histone lysine methylation state or IgG negative control (see Application Notes and Table 1).
Most popular related searches
pAG-MNase-mediated release or pAG-Tn5-mediated tagmentation of genomic chromatin and the barcoded nucleosomes is dependent on the specificity of the antibody used. After sequencing, the relative read count of each spike-in nucleosome barcode provides a quantitative metric of on- vs. off-target recovery (Figures 2 and 3) as well as quantitative sample normalization, thereby gauging experimental success, guiding troubleshooting efforts, and enabling reliable cross-sample comparisons.
Formulation: A mixture of 16 PTM-defined semi-synthetic nucleosomes conjugated to paramagnetic beads in 10 mM sodium cacodylate pH 7.5, 100 mM NaCl, 1 mM EDTA, 50% glycerol (w/v), 1x Protease Inhibitor Cocktail, 100 μg/mL BSA, 10 mM β-mercaptoethanol. Molarity = 3 nM each, 96 nM total nucleosome. Average MW = ~263,072.7 Da.
Storage and Stability: Store at -20°C. Lower temperatures can cause freezing and will permanently damage the magnetic beads. Stable for six months from date of receipt.
