

- Home
- Companies
- MtoZ Biolabs
- Services
- MtoZ Biolabs - Crosslinking Protein ...

MtoZ Biolabs - Model crosslinking-protein-interaction-analysis -Crosslinking Protein Interaction Analysis Service
Under physiological conditions, most protein interactions are transient, posing significant challenges for their study. Crosslinking reagents resolve this by covalently bonding interacting proteins, thereby capturing protein-protein complexes and preserving brief, weak interactions for easier separation and characterization. Researchers can perform crosslinking both in vivo and in vitro, choosing based on project needs. In vivo crosslinking maintains proteins in their native states, while in vitro crosslinking might lead to protein denaturation.
1. In Vivo Crosslinking
In vivo, crosslinking reactions take place within cells, reducing false positives and maintaining complex stability. Hydrophobic and lipophilic crosslinkers are used for proteins inside the cell membrane, whereas hydrophilic and water-soluble crosslinkers are preferred for membrane-surface proteins like plasma-anchored receptors. Despite proteins remaining in their native states, the complexity within cells makes optimization challenging. However, using crosslinkers with shorter arms can help mitigate non-specific crosslinking.
2. In Vitro Crosslinking
This method involves homogenizing and lysing cells, making it ideal for studying stable interactions. Lysing cells in specific buffers facilitates controlled reaction conditions such as temperature, pH, and ionic strength. The absence of in vivo environmental constraints allows for a broader selection of crosslinkers and better control of specificity.
Dülsner, E. et al. Nat Methods. 2009.
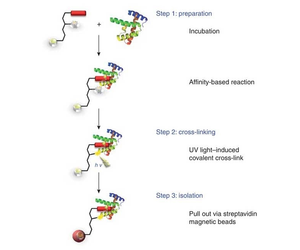
Figure 1. Analysis of Protein Interaction Using Cross-Linking Method
1. Chemical Crosslinking
Chemical crosslinkers covalently connect interacting proteins, structural domains, or peptides that are closely adjacent due to interactions, by forming chemical bonds between specific amino acid functional groups. Depending on the specific requirements of the project, many commercial chemical crosslinkers are available, including homobifunctional or heterobifunctional crosslinkers, long-arm or short-arm crosslinkers, cleavable or non-cleavable crosslinkers, water-soluble or non-water-soluble crosslinkers, etc. Homobifunctional molecules target the same functional groups on proteins, while heterobifunctional crosslinkers can target different functional groups on different proteins, allowing for greater variability or specificity in crosslinking. Crosslinker molecules can also be designed to include cleavable elements, such as esters or disulfide bonds, which can be reversed or disrupted by adding hydroxylamine or a reducing agent, respectively. Hydrophobic crosslinkers can enter hydrophobic protein structural domains or pass through cell membranes; Hydrophilic crosslinkers can interact well with the aqueous environment and promote the formation of crosslinked structures.
2. Photosensitive Crosslinking
To better analyze protein-protein interactions, more complex crosslinkers have been designed by adding photosensitive groups, etc. These photosensitive groups only react at selected times and only respond to ultraviolet light exposure. Heterobifunctional crosslinkers with a chemical crosslinking group on one end and a photosensitive group on the other can react with the selected target protein in two steps.
1.Analysis of Transient in Vivo Protein Interactions
2. Analysis of Low-Affinity in Vitro Protein Interactions
3. High Detection Sensitivity
1. Design of Crosslinking Methods and Types of Crosslinkers Based on Specific Project Requirements: In Vivo/In Vitro
2. Optimization of Reaction Conditions, Targeting a Combination of Sample Characteristics and Project Requirements, for Experimental Condition Optimization
3. Cell Lysis and Co-Immunoprecipitation (Co-IP)
4. Immunoblotting/Mass Spectrometry Analysis (as Determined by Project Requirements)
5. Analysis of Data and Delivery of Report
MtoZ Biolabs is an integrate contract research organization (CRO) providing advanced proteomics, metabolomics, bioinformatics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. The name of MtoZ represents “mass to charge ratio” in mass spectrometry analysis, as most of our services are provided based on our well-established mass spectrometry platforms. Our services allow for the rapid and efficient development of research projects, including protein analysis, proteomics, and metabolomics programs.
MtoZ Biolabs is specialized in quantitative multiplexed proteomics and metabolomics applications through the establishment of state-of-the-art mass spectrometry platforms, coupled with high-performance liquid chromatography technology. We are committed to developing efficient, and effective tools for addressing core bioinformatics problems. With a continuing focus on quality, MtoZ Biolabs is well equipped to help you with your needs in proteomics, metabolomics, bioinformatics, and biopharmaceutical research. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample.
Email: marketing@mtoz-biolabs.com
